General information
The following code serves as an example for running the spike induced fields (SIF) analysis.
In this example, SIFs are extracted from multi-electrode array data recorded using the multi-channel systems
(MCS - http://www.multichannelsystems.com/) MEA2100 amplifiers with an array. The dataset contains responses
Reading the data and meta data is peformed with the MCRackRecording class. The interface is specified in the
dataRecording class and can be modified for other formats when required. For data recorded with the MCRack software
by MCS MCRack should be downloaded and installed (http://www.multichannelsystems.com/software/mc-rack)
All analysis procedures are methods of the MEAAnalysis class.
Information about these recordings is provided by the MEARecordings excel table (see example)
The code uses two matlab packages:
Panel: https://de.mathworks.com/matlabcentral/fileexchange/20003-panel
GUI layout toolbox: https://de.mathworks.com/matlabcentral/fileexchange/47982-gui-layout-toolbox
Contents
Initiate object and set recording
create the MEAAnalysis object for the recordings in 'MEARecordings.xlsx'.
MA=MEAAnalysis('MEARecordings.xlsx');
MA.setCurrentRecording('recNames=AF16_250216_Looming3D0');
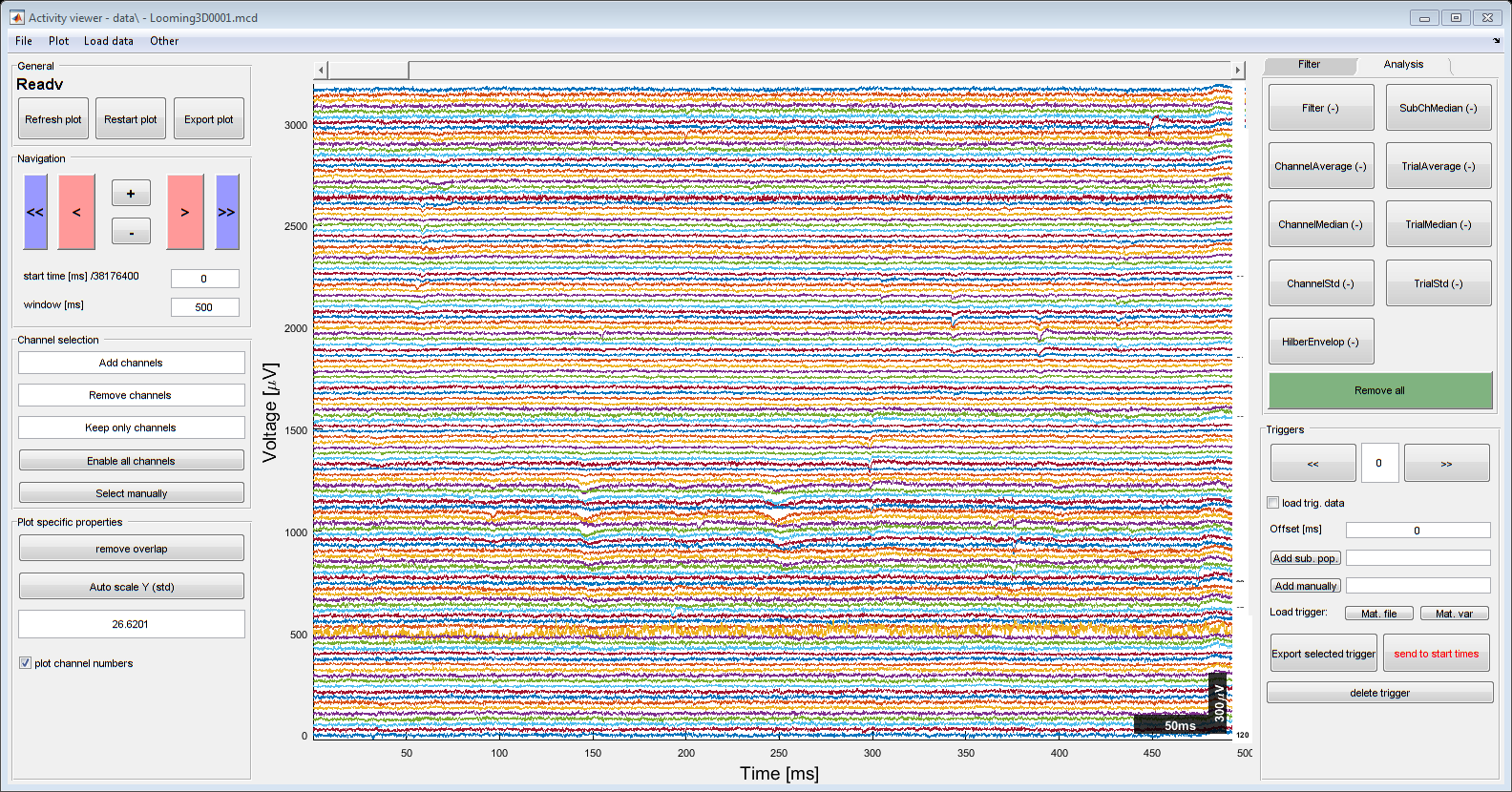
timeSeriesViewer(MA.currentDataObj);
Experiment data retrieved from: MEARecordings.xlsx
No .chMap files were found for this recording
Current exp. set to: Looming3D0*.mcd-Looming3D0*.mcd @ data
Filter initialized with sampling freq. of 1Hz
Spike sorting
Spike induced fields are extracted by averaging over spike triggered unfiltered raw data traces. Sorting can be perform
with any method. In this example, spikes were sorted with gridSorter (internally developed). The output of sorting
is located in the folder "Looming3D0001_spikeSort". To get the sorting with the grid sorter (may take one day of processing),
run "MA.getSpikeSorting;". Alternatively, if other sorted data exists, two files should be places in the spike sorting folder:
1) spikeSorting.mat - cotaining spike times for every neuron (t, ic)
2) STWaveform.mat - containing the average spike triggered waveforms (avgHPWF, avgRawWF, nSpkTotal, neuronNames)
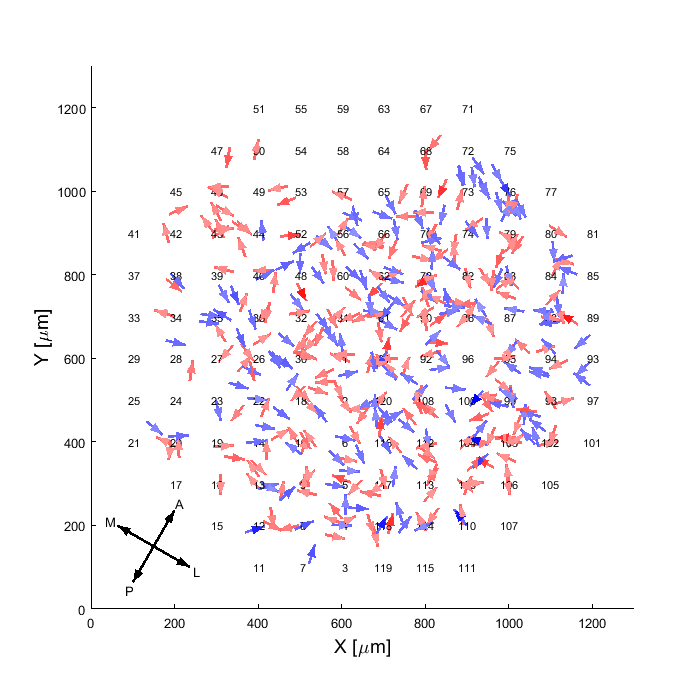
Triangulate spike position
Calculate the estimated position of the soma from the extracellular data. If this analysis was already performed and needs to be
overwriten run with property 'overwrite' set to 1: MA.getSpikePositionEstimation('overwrite',1);
To see the list of properties that can be modified for this method (or other methods), run MA.getSpikePositionEstimation('inputParams',1);MA.getSpikePositionEstimation;
Analysis results already exist for this method, use overwrite if needed
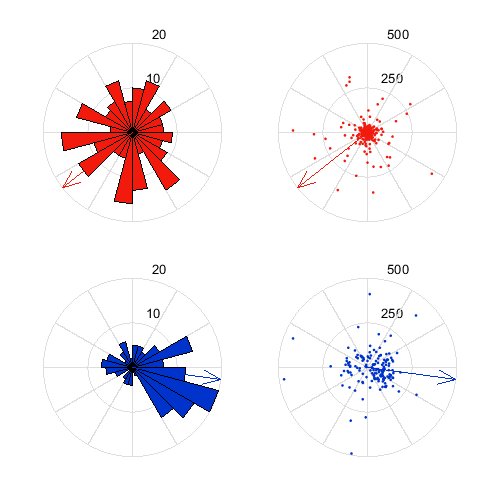
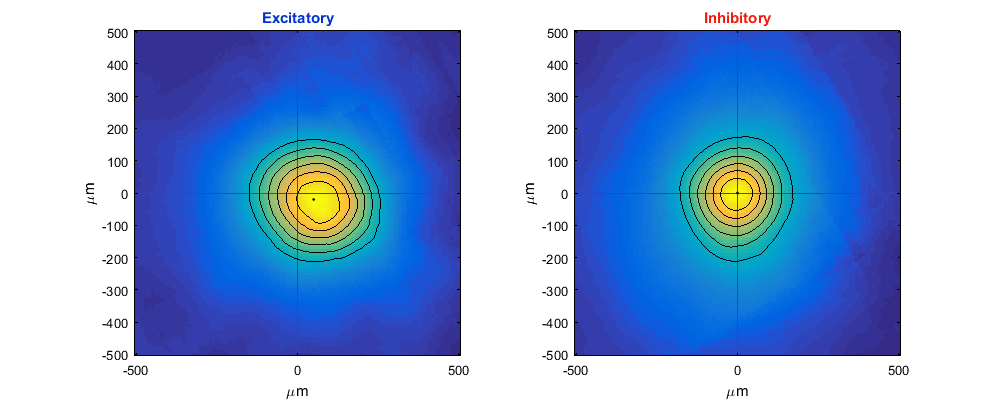
calculate and plot spike induced fields
MA.getSpikeInducedFields;
reverseDirction=MA.par.ctxFlip{MA.currentPRec};
angle2LateralRight=MA.par.ctxRotation{MA.currentPRec};
MA.plotSpikeInducedFields('angle2LateralRight',angle2LateralRight,'reverseDirction',reverseDirction);
MA.plotAvgSIFMap('angle2LateralRight',angle2LateralRight,'reverseDirction',reverseDirction);
spike induced fields analysis already exists. use overwrite to run again
Getting positions from layout files
Getting positions from layout files